Guest Article: Beauty and the Building Blocks…
Jun0

The building blocks of life. Photograph courtesy of Dr. Ron Zuckermann
…a nanoscientist’s quest to mimic Nature’s molecular blueprints
Have you ever found yourself entranced by the exquisite beauty and complexity of living things? Like the intricacies of a budding flower, or the mesmerizing patterns on a butterfly’s wing? Have you ever wondered: “what are living things made of?” Are these materials just as beautiful if we were to zoom way way in and look at the actual molecular building blocks that make up life? Take a look at the interactive link The Scale of Things to see just how small the building blocks of life really are! Well the answer is “OMG – totally!” All living things share a ubiquitous set of molecular building materials we call proteins, and they are absolutely stunning! They are not only smashingly beautiful to look at, they are capable of performing a mind-numbing myriad of very intricate and complex functions that are essential to life. In a very special guest post, leading nanoscience Professor Ron Zuckermann of the renowned Lawrence Berkeley National Laboratory recounts his life’s mission as a chemist to try and build artificial microscale sheets made up of nature’s very own building blocks—proteins. Everything you wanted to know about what nanotechnology is, exactly, why engineering proteins is the science of the future, and what we plan to use these discoveries for, under the “continue reading” cut.
I am a Materials Scientist working in a nanoscience research institute called The Molecular Foundry. A fundamental problem in nanoscience is how to make man-made materials with a similar level of precision and complexity at the molecular level found in nature. I am interested in applying lessons from the world of protein structure to practical man-made materials. If we are successful, we should be able to make materials that are cheap and rugged like a piece of plastic, yet be able to perform highly sophisticated functions, like recognizing a molecular partner with high specificity, or even catalyzing chemical transformations. Such materials could be used to make sensors for the detection of harmful chemicals, or as robust medical diagnostics that could survive harsh conditions, say in an underdeveloped nation. In a nutshell, we aim to make artificial proteins. This is an incredibly difficult problem, and one I have been working on for more than 20 years now. Sound a bit ambitious, or maybe a bit crazy? As I will describe in this article, it may actually be quite possible if we break the problem down into bite size chunks. The challenge comes down to two fundamental things: design and synthesis.

The beauty of proteins on a nano scale: 3-Dimensional structures of two famous proteins showing the intricate fold of the polymer chain. Green fluorescent protein (left) is mostly comprised of flat ribbons called beta-sheets, and myoglobin (right) consists primarily of repetitive loops called alpha-helices.
Protein architecture
When I look at the molecular structure of a protein molecule, I see an architectural blueprint that has survived untold generations of evolution and optimization. How then do we break open the hidden rules in these structures and use them to guide us in the design of man-made materials? Over the past several decades, scientists have used the biophysics techniques of X-ray crystallography and nuclear magnetic resonance spectroscopy (NMR) to determine the exact three-dimensional (3D) structure of thousands upon thousands of proteins. What’s cool is that these are all available for anyone to look at (for free!) and study in the Protein Data Bank.
The most fundamental thing to notice is that nearly all protein structures share the following characteristics: (1) they are made of a linear polymer chain that is folded into a precise 3D structure and (2) they are comprised of only 20 simple molecular building blocks called amino acids, arranged in an exact sequence along the polymer chain. When we think of a ‘polymer’ we think of a long chain of repeating chemical building blocks (called monomers) found in materials like nylon or polyethylene. Such man-made materials are incredibly useful and ubiquitous now in our environment (plastic bags or saran wrap, for example). But nature beat us to the punch a long, long time ago. Biopolymers, like proteins and nucleic acids are fundamentally way more sophisticated than man-made polymers. Even though they share the same basic architecture – a linear chain of chemical building blocks – biopolymers contain information encoded in their monomer sequence. This is not unlike the way we store information in a computer. But instead of a long string of 1’s and 0’s, nature uses long polymer chains of either 4 nucleotides (the building block units of RNA and DNA), or 20 amino acids (the building block units of proteins). These 20 chemically distinct amino acid building blocks are arranged in a particular order along the chain that we refer to as the protein’s “sequence.” This sequence is powerful because in many cases it provides all the information or “molecular instructions” necessary for the polymer chain to fold up into a precisely defined 3-dimensional structure. Once folded, the protein is poised and ready for action. The fields of Structural Biology and Protein Folding have revealed the exact way that proteins fold to form local “secondary” structures, called alpha helices and beta sheets, and how these assemble together to create the fully folded protein structure. Think all this sounds a bit too complicate? Try visiting FoldIt, a really fun video game where you can actually learn all about protein folding!
Protein Mimicry
If we ever hope to create man-made protein-like materials, it is safe to assume that we will need a polymer system that shares some of the basic protein-like characteristics: for example, they will need to have a sizable set of chemically diverse monomer building blocks that can be arranged in a particular order along a linear polymer chain of at least 50 monomers long. This is a quite a tall order simply from a chemical synthesis perspective. Moreover, once we are over that hurdle, design tools will be needed to help us figure out which sequences to make.
In the early 1990s, I invented a way to synthesize a new family of non-natural polymers we called “peptoids.” I had just graduated from UC Berkeley with a PhD in organic chemistry and joined a start-up biotechnology company to develop new technologies to accelerate drug discovery. We developed peptoids to be potential therapeutic drugs. The cool thing about peptoids is that the building blocks are very very close in structure to Nature’s amino acids, but different enough to be much more rugged. They can be made from very cheap and simple chemical building blocks, and they can be made in any predetermined sequence you want. We soon developed robotic synthesizers to automatically synthesize these materials for us (see below).

Robots march into the chemistry lab. Robots are used to automatically synthesize protein-like polymers.
Before long we discovered that short peptoid chains (just 3 monomers long) could have potent biological activities and showed promise as drug candidates. But what really floored me was that the peptoid synthesis chemistry we developed worked so efficiently that we could link over 50 monomers together, one after the other. This means each building block was being attached to a growing chain with an incredible accuracy of over 99.5%! This was exactly the tool we needed to begin the quest for creating artificial proteins. We had discovered the most efficient and practical way to make non-natural polymers of a specific sequence. This was completely awesome!
There was only one problem – the company I worked for had no interest in such a bold quest into basic science. How could such a pursuit lead to a moneymaking product in a few months? In 2006 I moved to Lawrence Berkeley National Laboratory where I set out in earnest to search for artificial proteins. Fortunately, tackling difficult problems in basic science is much more the norm here. And more good news – my previous employer was kind enough to donate my robots to me. Armed with this technology to synthesize peptoid polymers, we turned to the next daunting task: which sequence of monomers should we make? It turns out that there are an absolutely astronomical number of possible peptoid sequences that can be made. Consider that there are several hundred building blocks to choose from at each of the 50 positions of a polymer chain. This means there are over 100 to the 50th power potential sequences to choose from. This is more than the number of atoms in the universe! What was a chemist to do?
Like Oil and Water
To help us focus our sequence design, we once again turned to nature. A long-time collaborator and friend of mine, Professor Ken Dill of UC San Francisco has studied protein structure in detail for decades, and has distilled some fundamental rules that are universal to all protein structures. He notes that protein structures are like globes with a water-loving surface and a water-hating (or oil-like) interior. The bottom line is you can basically lump each amino acid in the protein’s sequence into one of two categories: oil-like or water-like. This simple classification can tell you whether an amino acid is located on the inside or the outside of the protein.
The amazing thing about this insight is that it’s like looking at a protein wearing X-ray glasses! Instead of seeing 20 different “colors” of amino acids, we see only two: black and white. We are back to a simple binary code—like 1’s and 0’s. This makes it much easier to see the secret patterns hidden within the sequence. In fact, many researches have convincingly demonstrated that these binary patterns are simple, plentiful and predictable.
So with our handy X-ray glasses on we returned our gaze to the peptoid structure problem. We realized that all our sequence recipe needed was a touch of Dill! This meant we could greatly simplify our search for the right peptoid sequence. We needed to only consider two, diametrically opposed building blocks: water loving and water hating.
Nanosheets
Inspired by these insights, we set out to find the right sequence patterns that would result in a precisely ordered structure in a non-natural peptoid polymer. We began to systematically unlock the sequence code by using our robots to synthesize all the possible repeating patterns of these two disparate building blocks. We reasoned that if we were to make something precisely ordered, it would “crystallize” into something that we could see. Fortunately we have really powerful electron microscopes in my building. So we started cranking through all the possible sequence patterns, dissolving up each new sequence in some water and taking it down to the basement to look at them really close up.
Now it is a little nuts to think that we could make something precisely structured from a peptoid polymer, since it is known that each strand is about as stiff as a piece of overcooked spaghetti. And as one might expect, most of our sequences looked really messy and gooey. But very soon we saw something quite striking. In one particular sample, we saw large, flat paper-like objects with sharp, straight edges. And they were floating around everywhere in the solution. An unexpected sight to be sure!
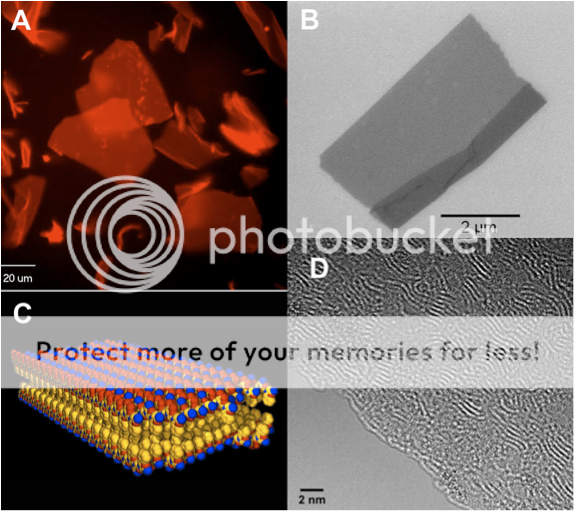
Ultra-thin nanosheets with protein-like structures form spontaneously in water. The peptoid nanosheets sheets: (A) are floating around in solution as observed by optical microscopy, (B) have very sharp edges as observed by scanning electron microscopy, (C) are only two molecules thick, and (D) individual polymer chains can be directly observed by abberration-corrected transmission electron microscopy.
Fast-forward another year of making careful measurements and reproducing the results over and over. We determined that these sheets were only two molecules thick, and yet millions and millions of molecules wide. We had discovered the largest and thinnest organic crystals ever! We were able to use one of the most powerful electron microscopes in the world in the National Center of Electron Microscopy, to look directly at the individual polymer chains that make up the crystal. We could see these chains wiggle around and slide against one another as if they were alive. No one had ever seen such detail before – a truly awe-inspiring sight!
We were able to use many kinds of advanced analytical tools to tell us that these nanosheets were indeed very special. They have the same kind of ordered structure that a protein has: they have a defined inside and outside, and we know almost exactly where each atom is located in the structure. Essentially, we discovered the sequence code that programs the polymer chain to form a 2D sheet. No doubt there are more complex codes waiting to be discovered that will form even more sophisticated structures. This discovery was recently reported in the journal Nature Materials, and was picked up by the more mainstream publications WIRED.com and Chemical & Engineering News. These sheets are likely to be important for all kinds of potential applications. Their discovery is kind of like the invention of ‘molecular plywood’: a new kind of nanoscale building material from which we can engineer even more complex molecular architectures. It’s amazing what kind of beauty you can create from simple building blocks!
Basic research like this can move seemingly very slowly, which makes the occasional breakthrough like this all the more meaningful and exciting. It reaffirms for me that it is so important to listen to and cultivate your inner curiosity, surround yourself with like-minded people, and aim high. With enough patience and persistence, wonderful things await discovery!
Take a look at a brief video of Dr. Zuckermann explaining his lab’s nanosheet discovery:
Ron Zuckermann is the Facility Director of the Biological Nanostructures Facility at Lawrence Berkeley Laboratory. Dr. Zuckerman also provides numerous consulting services at the intersection of chemistry, biology and engineering.
*****************
ScriptPhD.com covers science and technology in entertainment, media and advertising. Hire our consulting company for creative content development.
Follow us on Twitter and our Facebook fan page. Subscribe to free email notifications of new posts on our home page.
No Comments »
No comments yet.